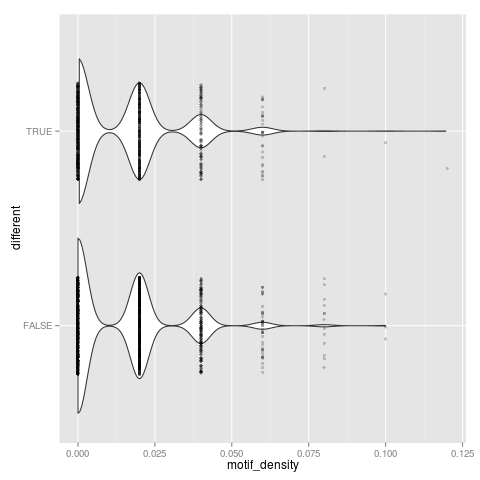
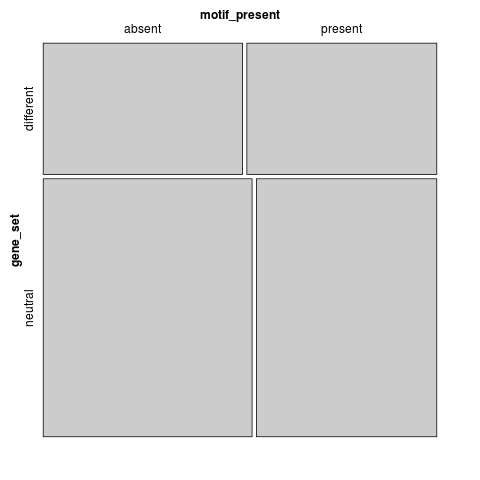
t-test: p=0.315957
Neutral set average density: 0.0128315
Different set average density: 0.0134809
motif_present gene_set absent present different 509 485 neutral 1048 905
Fisher's exact test: p=0.211879
Binomial GLM: p=0.207155
Call:
glm(formula = motif_present ~ different, family = binomial, data = daf)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.157 -1.116 -1.116 1.240 1.240
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.14670 0.04538 -3.233 0.00123 **
differentTRUE 0.09840 0.07801 1.261 0.20715
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 4075.9 on 2946 degrees of freedom
Residual deviance: 4074.4 on 2945 degrees of freedom
AIC: 4078.4
Number of Fisher Scoring iterations: 3