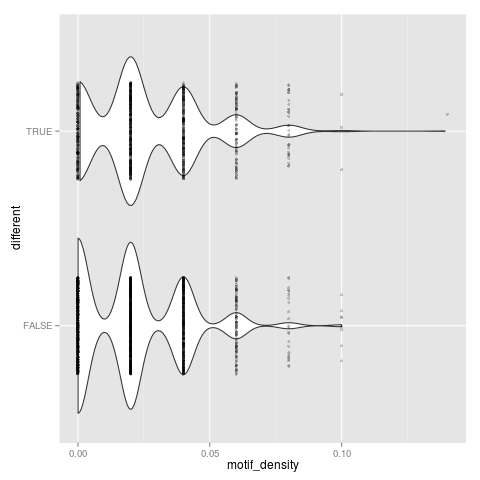
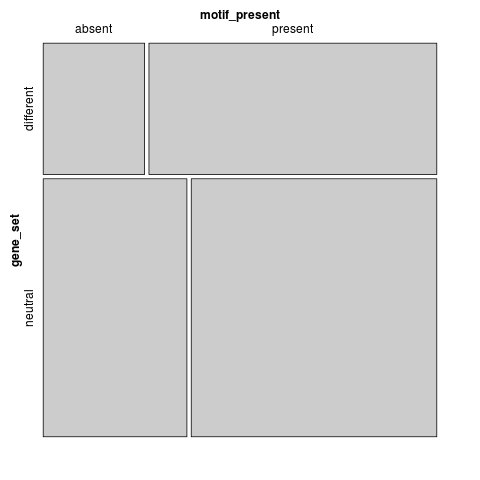
t-test: p=7.36949e-10
Neutral set average density: 0.0200512
Different set average density: 0.0250704
motif_present gene_set absent present different 259 735 neutral 721 1232
Fisher's exact test: p=2.46496e-09
Binomial GLM: p=3.88379e-09
Call:
glm(formula = motif_present ~ different, family = binomial, data = daf)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.6401 -1.4117 0.7770 0.9599 0.9599
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.53576 0.04689 11.426 < 2e-16 ***
differentTRUE 0.50729 0.08614 5.889 3.88e-09 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 3748.4 on 2946 degrees of freedom
Residual deviance: 3712.6 on 2945 degrees of freedom
AIC: 3716.6
Number of Fisher Scoring iterations: 4