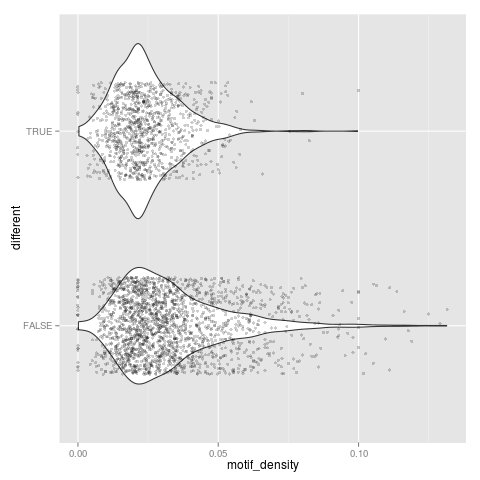
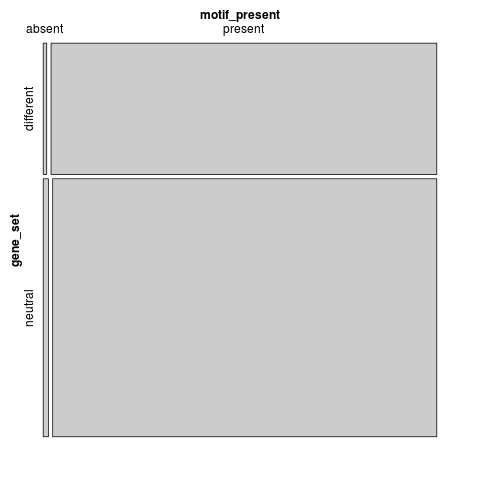
t-test: p=3.28326e-51
Neutral set average density: 0.0330517
Different set average density: 0.0242702
motif_present gene_set absent present different 9 985 neutral 26 1927
Fisher's exact test: p=0.371267
Binomial GLM: p=0.315975
Call:
glm(formula = motif_present ~ different, family = binomial, data = daf)
Deviance Residuals:
Min 1Q Median 3Q Max
-3.0674 0.1349 0.1637 0.1637 0.1637
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 4.3056 0.1974 21.808 <2e-16 ***
differentTRUE 0.3898 0.3887 1.003 0.316
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 379.91 on 2946 degrees of freedom
Residual deviance: 378.84 on 2945 degrees of freedom
AIC: 382.84
Number of Fisher Scoring iterations: 7
Binomial GLM adjusted for length: p=0.169625
Call:
glm(formula = motif_present ~ length + different, family = binomial,
data = daf)
Deviance Residuals:
Min 1Q Median 3Q Max
-3.4530 0.0420 0.1174 0.1954 0.4165
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.672434 0.332742 8.032 9.63e-16 ***
length 0.014479 0.003261 4.441 8.97e-06 ***
differentTRUE -0.560586 0.408171 -1.373 0.17
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 379.91 on 2946 degrees of freedom
Residual deviance: 339.16 on 2944 degrees of freedom
AIC: 345.16
Number of Fisher Scoring iterations: 9